Using cryo-EM SPA (single particle analysis) technology, Shuimu Biosciences has been able to resolve the three-dimensional conformation of CD73 dimer and full-length antibody complexes, elucidate the non-competitive mechanism of action of antibodies based on spatial epitope information, and visualize the differences between them and similar antibodies, providing underlying evidence to support eye-catching efficacy data in clinical trials.
Introduction
In May, Nature Review Drug Discovery published a review "FDA approves 100th monoclonal antibody product" [1], and the number of approved drugs in the past decade shows that antibody drug development has almost skipped the process from 0 to 1 and entered an explosive growth. The number of approved drugs in the last decade shows that antibody development has almost skipped the process from 0 to 1 and has gone straight to explosive growth. The fast-growing market has also given rise to a highly competitive situation with several or even dozens of investigational/clinical drugs for a single target, forcing pharmaceutical companies to seek new and different drugs, but at a time when clinical trial control groups are becoming more stringent and clinical value has a veto power, innovation and differences also mean great risks.
How can we get more information in the early stages of antibody development and strengthen the evidence chain to reduce the risk of clinical trial failure? How to trace the mechanism of action of a drug based on the validation of in vitro and in vivo trials? How can we "see" the actual difference in the product after the innovation with the most visual evidence while boasting the innovation of the technology platform? Structure-based spatial epitope information may provide the answer.
There is no approved CD73 drug on the market yet, but this year an antibody to CD73 was selected as one of the "Top 12" studies in the AACR [2], and Shuimu BioSciences has the honor to undertake the structural analysis of this antibody drug. In this paper, we will use this project to explain the unique antigen-antibody binding mode to avoid the hook effect of antibodies, and extend it to the identification of spatial epitopes based on Cryo-EM technology to enable the development of differentiated antibodies.
CD73: A Rising Star Target for Antitumor Drug Development
Immune detection site inhibitors represented by PD-1 and CTLA-4 have shown "long trailing" overall survival advantage in pan-cancer therapy [3] and are gradually extending from advanced tumor therapy to early adjuvant/neoadjuvant therapy [4], but the problem of low response rate is still a major obstacle to clinical application [5]. With the combination of unmet clinical needs and profit-driven factors, it is not difficult to understand the behavior of pharmaceutical companies to bury their heads in the hard search for new targets for immunotherapy. In addition to the tumor itself such as the number of neoantigens, host causes, especially the state of the local immune microenvironment of the tumor, are closely related to the efficacy of immunotherapy, and how to lift the inhibitory effect of the tumor microenvironment on immune cells and make cold tumors hot becomes a major strategy to improve the therapeutic effect rate and overcome immunotherapy resistance [6].
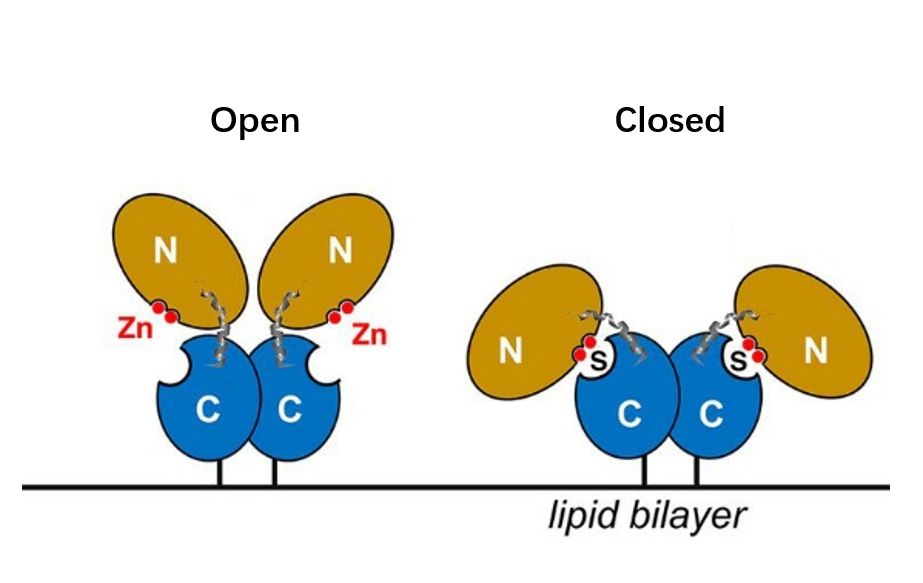
One of the important mechanisms in the formation of tumor suppressive immune microenvironment is the activation of adenosine production pathway. Adenosine triphosphate ATP and adenosine ADO are the same signals that regulate tumor cell growth and immune cell status, but unlike the pro-inflammatory effect of ATP, its metabolite ADO can activate G protein-coupled receptors A2A and A2B, which can inhibit the proliferation and cytotoxic effect of T cells and block the dendritic cells by inhibiting by inhibiting the proliferation and cytotoxic effects of T cells and blocking the differentiation and maturation of dendritic cells, which put immune cells in the tumor microenvironment in a hyporesponsive state (Figure 1), and also based on this, hypoadenosine has become an important idea for antitumor drug development [7].
One of the pathways of ADO production in vivo is the hydrolysis of ATP by the extracellular nucleotidases CD39 and CD73 in sequence to produce ADO (Figure 2, red line marked part), and the development of inhibitory small molecules or antibody-based drugs targeting CD73 can block the conversion of ATP to ADO [8].
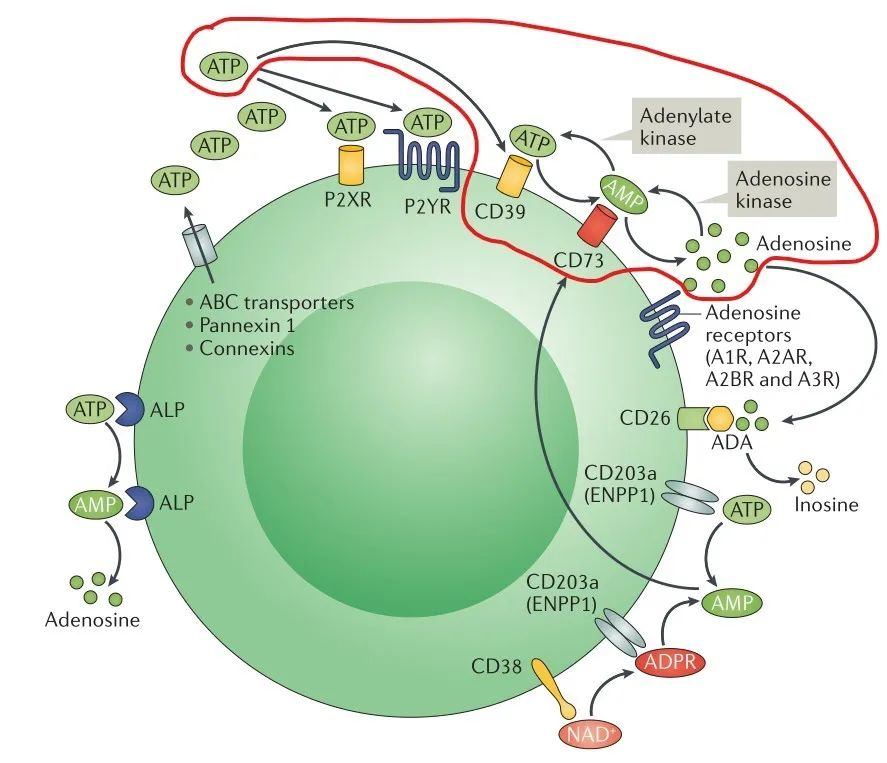
CD73, also known as extracellular 5'-Nucleotidase (Ecto-5'-Nucleotidase), is responsible for the further dephosphorylation of AMP to ADO after ATP is catalyzed by CD39 to produce adenosine monophosphate AMP. CD73 exists in vivo as a non-covalent dimer, and CD73 The N-terminal and C-terminal structural domains of the monomer are connected by a short α-helix, and structural biology studies as early as 2012 captured two distinct conformational states of CD73 - the open and closed conformations - with relative shifts of up to 114° between the N-terminal and C-terminal domains in both states (Movie 1). The gray sphere in the N-terminal domain marks the metal-zinc ion binding site, while the substrate binding site is located in the C-terminal domain, and the red dendritic pattern represents the ligand; after the C-terminal domain binds the AMP substrate, the C-terminal and N-terminal domains are in close proximity in the closed state, and the substrate and metal ion binding site together form the catalytic center of the enzyme. The N-terminal/C-terminal structural domains are in close proximity to each other, and the interaction interface and spatial conformation of the enzyme determine the selectivity of the enzyme for catalytic substrate monophosphate nucleotides, while the larger rotation between the two structural domains facilitates the release of the hydrolysis product ADO [9].
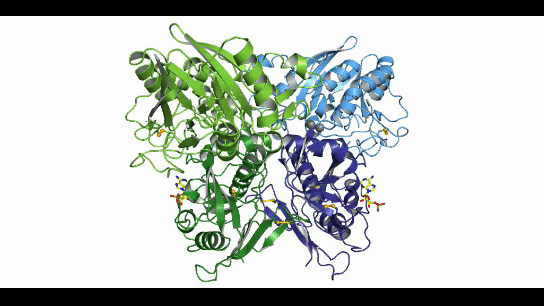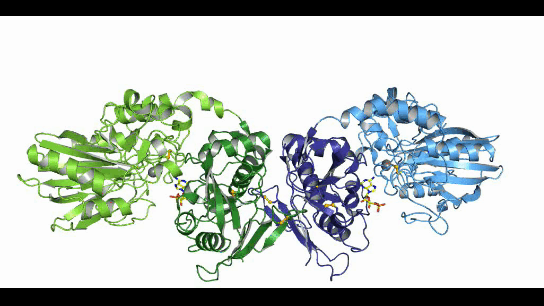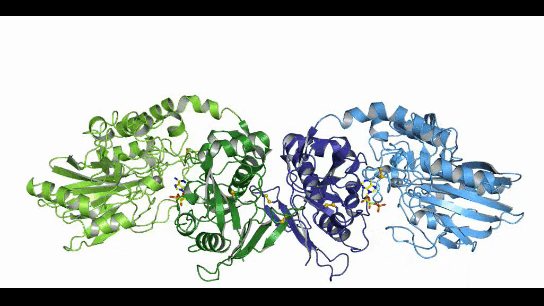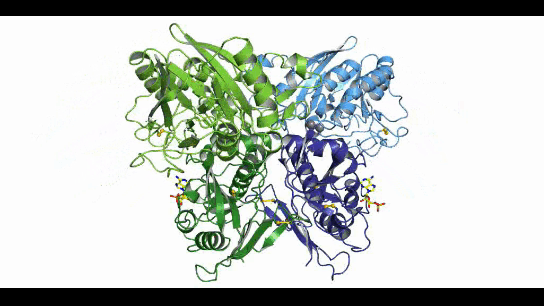
Movie 1. two conformations of human-derived CD73 dimer, Closed (activated) and Open (inactivated) [9]
Progress in structural studies of CD73 therapeutic antibodies
Although Sanofi has developed dual antibodies targeting different epitopes of CD73, only TB19 is analyzed because it mainly inhibits through TB19 and the binding epitope is more specific.
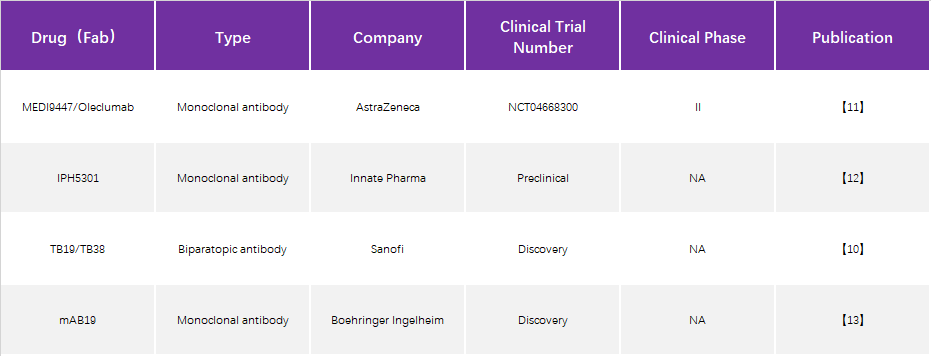
1. MEDI9447 (also known as Oleclumab)
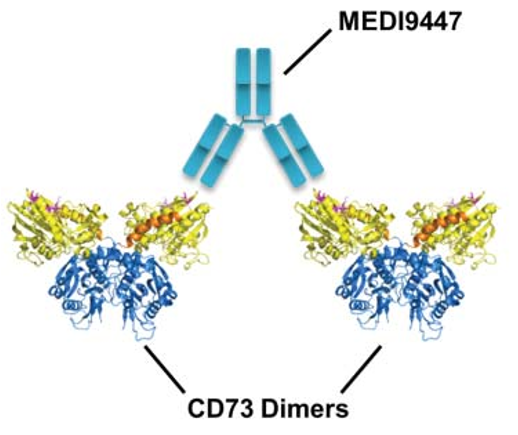
An antibody drug that was developed early and has received considerable attention. Using techniques such as hydrogen-deuterium exchange and site-directed mutagenesis, MEDI9947 is presumed to bind at the apical, outlying position of the N-terminal structural domain of CD73 antigen. As an allosteric inhibitor, antibody binding does not significantly alter the tertiary structure of CD73 protein (Figure 3).
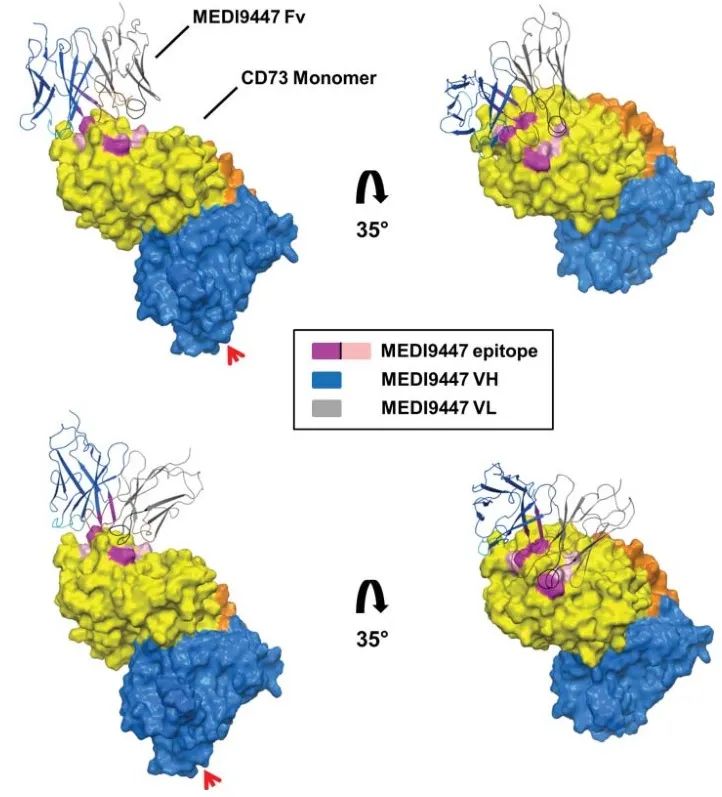
(Virtual docking using the crystal structure of the CD73 dimer in open conformation, with the three most important antigen-antibody interactions marked in rose and the three secondary antigenic epitopes in pink)
Based on the information on the epitopes bound by the antibody and the molecular weight of the antigen-antibody complex, it is hypothesized that the two Fabs of an antibody bind one of the two CD73 dimers, thereby holding the two CD73 dimers together (Figure 4) and blocking the catalytic activity by limiting the conformational change that activation must undergo. In this inter-dimer mode of action, both Fabs of the antibody must simultaneously bind to the N-terminal end of CD73 in order to inhibit enzymatic activity, which requires an extremely demanding antibody-antigen ratio. In practice, it is difficult to guarantee a 1:1 molar ratio of antibody to antigen in vivo, and if the ratio of antigen to antibody is not appropriate, the hook effect, which is a major concern in antibody drug development, will occur, i.e., when the concentration of the antigen CD73 is fixed, increasing the antibody dosage will result in a decrease in efficacy, and the hook effect was indeed observed in MEDI9947 drug development and clinical trials. effect was indeed observed in MEDI9947 drug development and clinical trials.
2. IPH5301
Although the resolved structure is the complex state of the single Fab of IPH5301 antibody bound to CD73, the mechanism of antibody action can be inferred by combining the negative staining results of the complex formed by the full-length antibody with CD73 and the orientation of the Fab in the structure of the Fab-CD73 complex, i.e., the two Fabs of the full-length antibody bind two monomers of the same CD73 dimer, because the antibody Because of the Y-configuration restriction of the antibody (the constant regions of the two Fabs are pulled together by the Fc), the antibody binds and holds the two N-terminal ends of the dimer in an intermediate state between Open and Closed. This model was also validated by an in vitro Fab inhibition enzyme activity assay - the addition of Fab alone did not inhibit CD73 catalytic activity, suggesting the importance of the full-length antibody Y conformation for CD73 activity inhibition. Because two Fabs of an antibody need to bind simultaneously to two monomers of the same dimer, it is also known as the intra-dimer binding mode. Although different from the inter-dimer binding mode of MEDI9447, it is speculated from the model of action that if the antibody is overloaded relative to the CD73 dimer, it is likely that only one Fab of the antibody binds to the CD73 monomer, so that the effect on the two N-terminal ends of the CD73 dimer is lost and there may be a risk of hook effect as well.
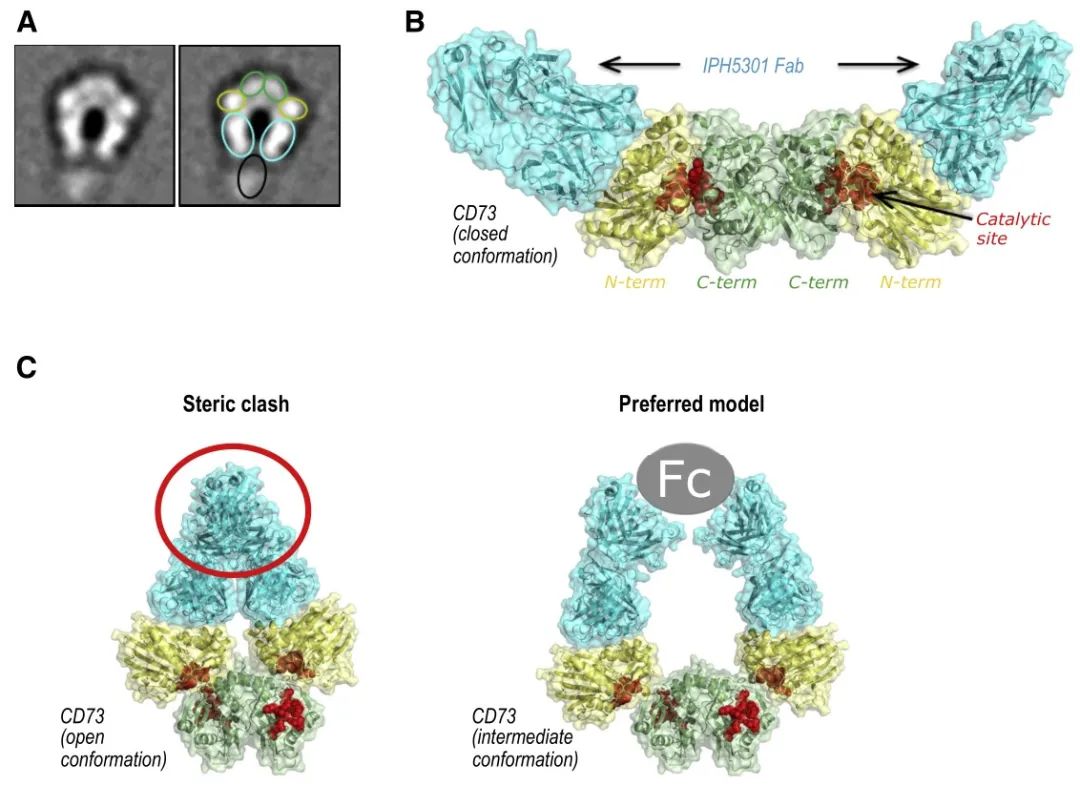
(A: representative negative staining conformation of CD73-IPH5301 full-length antibody complex, green box represents the N-terminal structural domain of CD73, yellow represents the C-terminal structural domain, green-blue represents the Fab of IPH5301, black represents the Fc of IPH5301; B: high-resolution structure of CD73 and IPH5301 Fab complex; C: on the left side is the binding of antibody Fab in the open state of CD73, and on the right side is the presumed binding mode of CD73 between Open and Closed. (the left side is the binding of antibody Fab in the open state of CD73, and the right side is the presumed binding mode, i.e. the antibody uses the Y conformation to fix CD73 in an intermediate state between Open and Closed, where the N-terminal and C-terminal cannot be closed to form the catalytic center)
3. TB19
TB19 also binds to the N-terminal end of CD73 monomer, but the antigenic epitope is clearly different from the above two antibodies, although the angle between the two structural domains of CD73 monomer after binding Fab is also in the intermediate state between Open and Closed, but the mechanism of action is completely different. The closed and open conformations are shifted by about 22 Å and 27 Å, respectively (Figure 6A), demonstrating that the N and C termini are "locked" in the non-activated intermediate state, but the spatial positions of the C-terminal substrate binding region and the N-terminal ion binding region are not close together, not because the steric conformation of the Y-type antibody This is not because the stereo conformation of the Y-type antibody restricts the opening angle of the two Fabs, but because the bound Fab occupies the position of the N-terminal domain in the active closed state (Figure 6B). Under this mechanism of action, the inhibitory effect of TB19 on CD73 does not depend on a specific bivalent binding mode, which theoretically reduces the probability of hook effect. However, TB19, as a monoclonal antibody, does not have a high affinity for CD73 and has strong inhibitory activity only at submicromolar concentrations, so TB19 is used as a parent for modifying bispecific antibodies and cooperates with other antibodies to achieve synergistic effects.
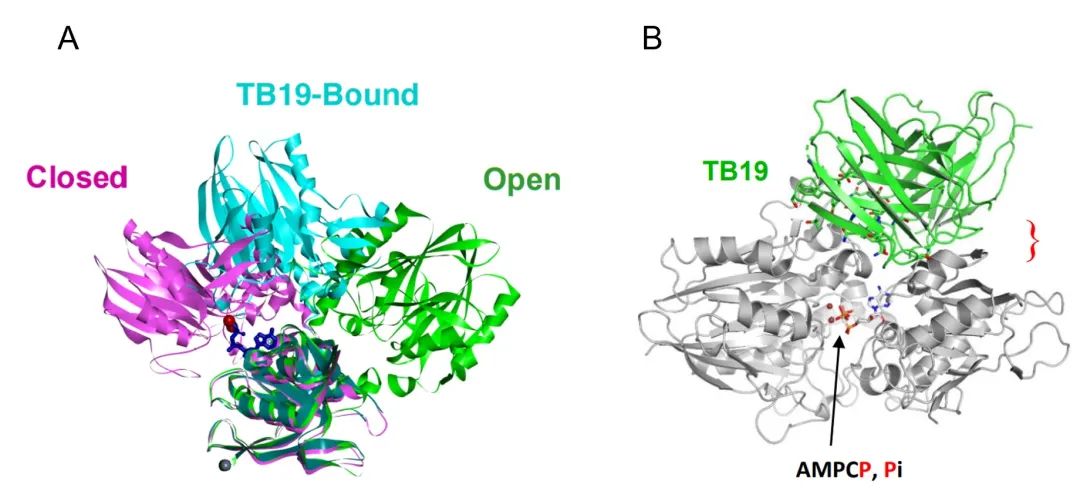
(A: The N-terminal and C-terminal ends of TB19 are in the intermediate state between Open and Closed after binding, and the zinc ion binding position is far from the substrate binding position; B: The published Closed conformation was fitted to determine the spatial resistance of TB19 and the N-terminal structural domain under the Closed conformation)
Project case structure dissection
While the previously mentioned antibodies either had to be in full-length to exert enzymatic activity inhibition or were ineffective as monoclonal antibodies requiring modification to dual antibodies for potentiation, the monoclonal antibody studied in this project can exert equivalent potency even if Fab is monovalently bound and does not rely on the Y-shaped structure of the antibody, suggesting that its mechanism of action may be different from any of the previously mentioned antibodies.
As the only antibody that binds to the C-terminal domain of CD73 protein, the antibody binds at the outer edge of the interface where the C-terminal domain forms a dimer, which is close to the substrate binding site, but does not prevent the substrate from binding to CD73 (Figure 7A). The most common mechanism of antibody action is antibody-substrate competition, but this not only requires high antibody antigen affinity, but the diversity of substrate binding also sets a barrier to antibody screening, and more importantly, the active regions of different enzyme-binding substrates often have high sequence homology, increasing the potential risk of drug off-target . Therefore, non-substrate competitive binding of antibodies is also an important idea for antibody drug development by locking the protein conformation in an inactivated state through an aliasing mechanism, thus exerting an enzymatic activity inhibition.
Comparing the structure of CD73 dimer bound to antibody Fab with the conformation of CD73 dimer in both Open and Closed states, it can be seen that the CD73 dimer obtained from this project is in an intermediate conformation between Open and Closed states, and the N-terminal structural domain of the antibody and the protein in the Closed active state is significantly site blocked, and the antibody The antibody binds and prevents the substrate and ion-binding region from approaching each other. This mode of action allows for monovalent binding of CD73 in the inactivated state through occupancy, and the ratio of CD73 to antibody in the system does not significantly affect the inhibitory effect of the antibody. Moreover, antibody binding at the outer edge of the C-terminal structural domain of CD73 protein also implies a wider binding angle and higher binding probability. The structural study well validates and explains the results of in vitro affinity, enzymatic activity, and hook effect assays.
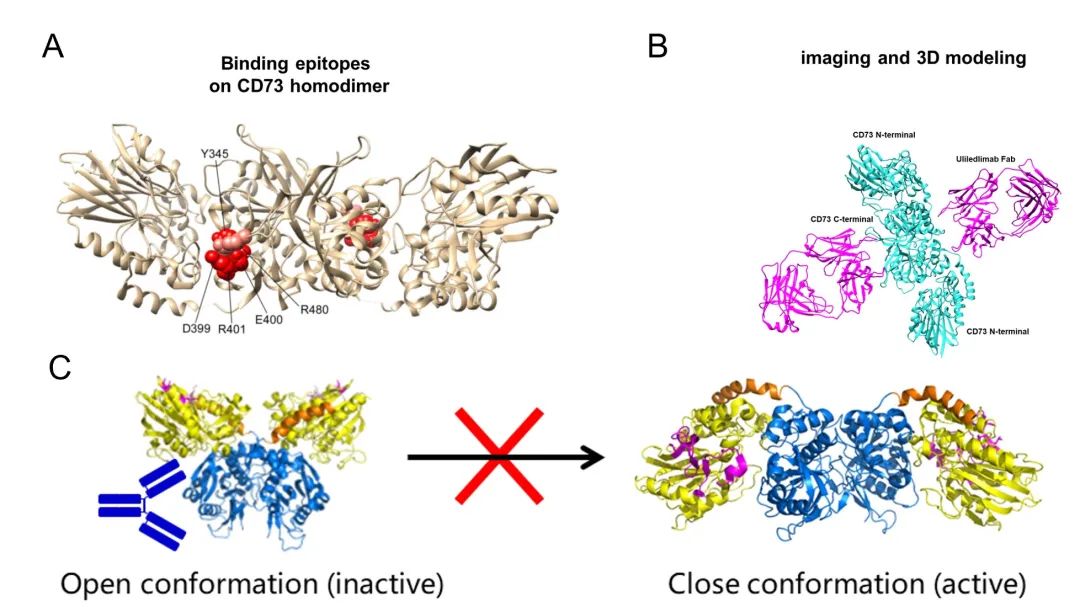
(A: atomic structure model of CD73 and key antigenic epitopes (marked in red); B: structural model of the final complex formed by the antibody Fab and CD73 dimer; C: spatial blockage between the antibody and the N-terminal structural domain of CD73, locking CD73 in the inactive state)
Spatial epitope identification technique based on cryo-EM technology
Cryo-EM high-resolution structures have become the standard for research papers in the field of neo-crown, and a whole set of systems on the evaluation of the potency of neo-crown neutralizing antibodies has been established based on spatial conformation and epitope information [14], but in fact, spatial epitope information is a very critical core data for the development of all large-molecule antibody drugs, which not only helps to understand the mechanism of action of antibodies and analyze conformational relationships, but even It may even be an important factor for antibody patent protection.
With the development of cryo-EM SPA (single-particle analysis) technology, more and more high-resolution structures of antigen-antibody complexes based on cryo-EM have been published [15-21]. Compared with other structural resolution techniques, the rapid popularity of cryo-EM in drug development lies not only in its ability to obtain conformations of biomolecules close to their physiological states, but also in its greater advantages in that.
1) No crystallization is required, and samples can be directly prepared for cryo-EM observation and structural analysis in the protein solution state, and for particles in good condition, structural analysis can be completed in as little as one day.
2) The total amount of protein required is as low as 0.1 mg, eliminating the time and cost of expressing a large amount of purified antibodies and antigens.
3) Cryo-EM has almost become the preferred means of analysis for difficult to crystallize proteins such as membrane proteins, proteins with large molecular weights, and proteins with extensive glycosylation modifications.
Conclusion
While pursuing surface differences such as smaller p-values in clinical trials, structure-based antibody epitope information can not only explain the mechanism of action of antibodies from the bottom, but also provide the most intuitive "differentiation". It also allows us to anticipate differences in the preclinical development phase and even evaluate and predict the performance of antibody candidates from a new perspective based on in vitro physiological and biochemical tests, making critical decisions more rational.
We hope that structural biology, represented by cryo-EM, will become the "wise eye" to help antibody drug development and help pharmaceutical companies to identify the most valuable antibodies from a wide range of antibodies. It is hoped that cryo-EM, represented by structural biology, will become a "wise eye" for antibody drug development, helping pharmaceutical companies to identify the most valuable strain from a wide range of antibodies.
Reference:
1. Mullard A. FDA approves 100th monoclonal antibody product[J]. Nature reviews. Drug Discovery, 2021.
2. Wang Z, Cao W, Zhang Y, et al. A differentiated CD73 blocking antibody with a unique intra-dimer binding mechanism for cancer immunotherapy[J]. 2021.
3. Garon E B, Hellmann M D, Rizvi N A, et al. Five-year overall survival for patients with advanced non‒small-cell lung cancer treated with pembrolizumab: results from the phase I KEYNOTE-001 study[J]. Journal of Clinical Oncology, 2019, 37(28): 2518.
4. Topalian S L, Taube J M, Pardoll D M. Neoadjuvant checkpoint blockade for cancer immunotherapy[J]. Science, 2020, 367(6477).
5. Schoenfeld A J, Hellmann M D. Acquired resistance to immune checkpoint inhibitors[J]. Cancer Cell, 2020, 37(4): 443-455.
6. Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies[J]. Nature reviews Drug discovery, 2019, 18(3): 197-218.
7. Vijayan D, Young A, Teng M W L, et al. Targeting immunosuppressive adenosine in cancer[J]. Nature Reviews Cancer, 2017, 17(12): 709-724.
8. Antonioli L, Yegutkin G G, Pacher P, et al. Anti-CD73 in cancer immunotherapy: awakening new opportunities[J]. Trends in cancer, 2016, 2(2): 95-109.
9. Knapp K, Zebisch M, Pippel J, et al. Crystal structure of the human ecto-5′-nucleotidase (CD73): insights into the regulation of purinergic signaling[J]. Structure, 2012, 20(12): 2161-2173.
10. Stefano J E, Lord D M, Zhou Y, et al. A highly potent CD73 biparatopic antibody blocks organization of the enzyme active site through dual mechanisms[J]. Journal of Biological Chemistry, 2020, 295(52): 18379-18389.
11. Geoghegan J C, Diedrich G, Lu X, et al. Inhibition of CD73 AMP hydrolysis by a therapeutic antibody with a dual, non-competitive mechanism of action[C]//MAbs. Taylor & Francis, 2016, 8(3): 454-467.
12. Perrot I, Michaud H A, Giraudon-Paoli M, et al. Blocking antibodies targeting the CD39/CD73 immunosuppressive pathway unleash immune responses in combination cancer therapies[J]. Cell reports, 2019, 27(8): 2411-2425. e9.
13. Wurm M, Schaaf O, Reutner K, et al. A novel antagonistic CD73 antibody for inhibition of the immunosuppressive adenosine pathway[J]. Molecular Cancer Therapeutics, 2021, 20(11): 2250-2261.
14. Barnes C O, Jette C A, Abernathy M E, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies[J]. Nature, 2020, 588(7839): 682-687.
15. Diwanji D, Trenker R, Thaker T M, et al. Structures of the HER2–HER3–NRG1β complex reveal a dynamic dimer interface[J]. Nature, 2021: 1-5.
16. Kumar A, Planchais C, Fronzes R, et al. Binding mechanisms of therapeutic antibodies to human CD20[J]. Science, 2020, 369(6505): 793-799.
17. Rougé L, Chiang N, Steffek M, et al. Structure of CD20 in complex with the therapeutic monoclonal antibody rituximab[J]. Science, 2020, 367(6483): 1224-1230.
18. Chin S M, Kimberlin C R, Roe-Zurz Z, et al. Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab[J]. Nature communications, 2018, 9(1): 1-13.
19. Neijssen J, Cardoso R M F, Chevalier K M, et al. Discovery of amivantamab (JNJ-61186372), a bispecific antibody targeting EGFR and MET[J]. Journal of Biological Chemistry, 2021, 296.
20. Zhang H, Huang C S, Yu X, et al. Cryo-EM structure of ABCG5/G8 in complex with modulating antibodies[J]. Communications biology, 2021, 4(1): 1-9.
21. Hao Y, Yu X, Bai Y, et al. Cryo-EM Structure of HER2-trastuzumab-pertuzumab complex[J]. Plos one, 2019, 14(5): e0216095.